Current research projects:
-
Collective motion and self-organization of living active matter
-
Growth dynamics of general living matter
-
Bacterial motility in complex environments
-
Population dynamics, pattern formation and cell-environment interactions in natural and synthetic multicellular systems
1. Collective motion and self-organization of living active matter
As a fast-growing and interdisciplinary field, active matter science studies systems consisting of units where energy is spent locally to generate mechanical work. Active matter includes all living organisms from cells to animals, biopolymers driven by molecular motors, and synthetic self-propelled materials. We have a long-term interest in understanding and controlling the spatiotemporal self-organization of biological active matter consisting of motile microorganisms.
-
Viscoelasticity enables self-organization of bacterial active matter in space and time
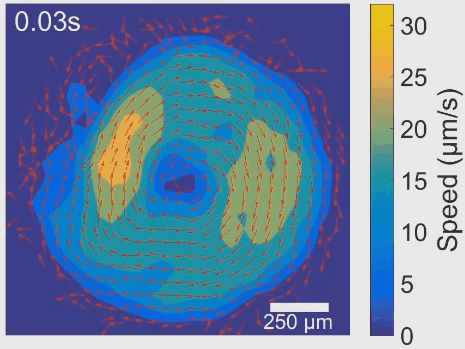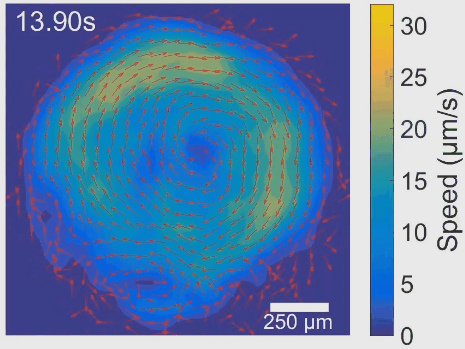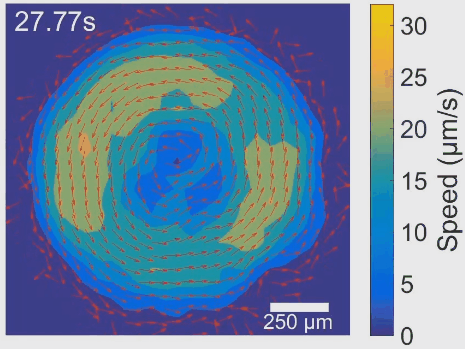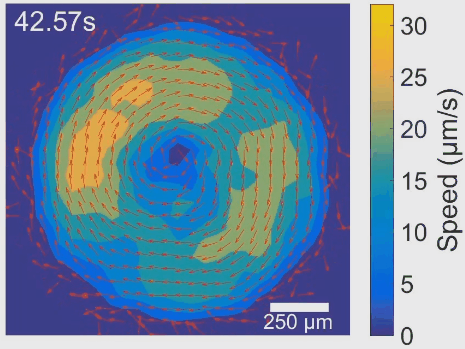
Simultaneous control of spatial and temporal order in active matter systems is challenging. To achieve this goal, we have found clues in viscoelasticity, a common property of complex fluids as having both fluid-like and solid-like responses under deformation. While manipulating the viscoelasticity of a bacterial active fluid with DNA polymers, we found spectacular phenomena: The bacterial active fluid first self-organizes in space into a millimeter-scale rotating vortex; then displays temporal organization as the giant vortex switches its global chirality periodically with tunable frequency, reminiscent of a self-driven torsional pendulum. The striking phenomena presumably arise from the interplay between active forcing and viscoelastic stress relaxation. This idea is supported by an active fluids model developed by our collaborators Cristina Marchetti (UC Santa Barbara) and Suraj Shankar (Harvard). In particular, linear stability analysis of the model shows that the onset of temporal order is triggered by flow instability via Hopf bifurcation. The discovery demonstrates experimentally for the first time that rheological properties can be harnessed to control active matter self-organization. It may pave the way for fabricating a new class of adaptive self-driven devices or materials, and has implications for the collective motion pattern of bacteria residing in biofilms and animal gastrointestinal tracts. See [Movie 1], [Movie 2].
References:
- Song Liu*, Suraj Shankar*, Cristina M. Marchetti & Yilin Wu (2021) Viscoelastic control of spatiotemporal order in bacterial active matter. Nature, 590 (7844), 80-84. (*co-first authors).
-
Collective oscillation without inherent local oscillators
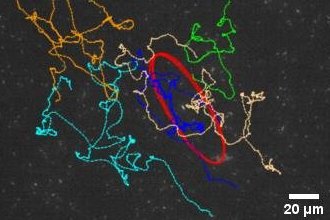
Collective oscillatory behavior is ubiquitous in nature and it plays a vital role in many living processes. Collective oscillations in multicellular systems studied to date often arise from coupling between individual cells that display inherent oscillations. In stark contrast, our lab discovered a new form of collective oscillation that does not require inherent oscillation of individual units: Millions of motile cells in dense bacterial suspensions can self-organize into collective oscillatory motion with robust temporal order, while individuals move in an erratic manner. We used a series of experiments to demonstrate that the phenomenon results from spontaneous symmetry breaking mediated by purely local interactions between individual cells. This ‘weak synchronization’ phenomenon presents a novel mechanism of oscillatory behavior in multicellular systems and constitutes a new type of ordered active matter. See [Movie 1], [Movie 2].
References:
- Chong Chen*, Song Liu*, Xiaqing Shi, Hugues Chaté, Yilin Wu (2017) Weak synchronization and large-scale collective oscillation in dense bacterial suspensions. Nature, 542, 210-214. (*co-first authors)
2. Bacterial motility in complex environments
-
Cohesive swimming of bacteria in two-dimensional confinement
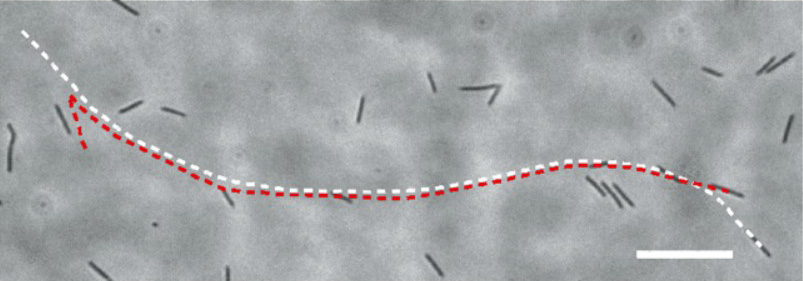
Characterizing bacterial interactions in 2D confinement will help to understand diverse microbial processes, such as bacterial swarming and biofilm formation. We discovered a novel form of interaction between flagellated bacteria in 2D confinement: When two or multiple nearby cells align their moving directions, they tend to swim side-by-side cohesively without direct cell body contact, as a result of short-range hydrodynamic interaction. We further found that cells in cohesive swimming move with higher directional persistence, which can increase the effective diffusivity of cells by ~3 times as predicted by computational modeling. The higher directional persistence may promote bacterial dispersal in unsaturated soils and in interstitial space during infections. See [Movie 1], [Movie 2], [Movie 3],
References:
- Ye Li*, He Zhai*, Sandra Sanchez, Daniel B. Kearns, Yilin Wu. (2017) Non-contact cohesive swimming of bacteria in two-dimensional liquid films. Phys. Rev. Lett. 119, 018101. (*co-first authors)
3. Population dynamics, pattern formation and cell-environment interactions in natural and synthetic multicellular systems
-
Dynamic motility selection promotes population segregation and collective drug tolerance in a bacterial swarm
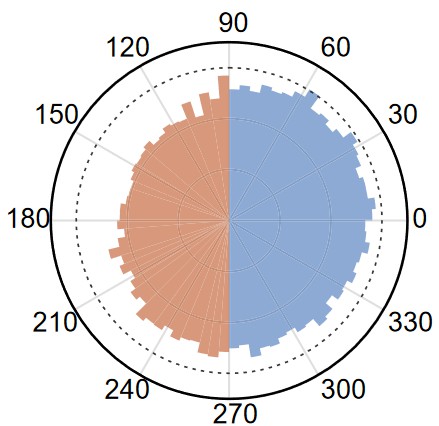
Population expansion in space, or range expansion, is widespread in nature and in clinical settings. Space competition among heterogeneous subpopulations during range expansion is essential to population ecology, and it may involve the interplay of multiple factors, primarily growth and motility of individuals. Structured microbial communities provide model systems to study space competition during range expansion. We used bacterial swarms to investigate how single-cell motility contributes to space competition among heterogeneous bacterial populations during range expansion. Our results revealed that motility heterogeneity can promote the spatial segregation of subpopulations via a dynamic motility selection process. The dynamic motility selection is enabled by speed-dependent persistence time bias of single-cell motion, which presumably arises from physical interaction between cells in a densely packed swarm. We further showed that the dynamic motility selection may contribute to collective drug tolerance of swarming colonies by segregating subpopulations with transient drug tolerance to the colony edge. Our results illustrate that motility heterogeneity, or “motility fitness,” can play a greater role than growth rate fitness in determining the short-term spatial structure of expanding populations.
References:
- Wenlong Zuo & Yilin Wu (2020) Dynamic motility selection drives population segregation in a bacterial swarm. Proc. Natl. Acad. Sci. USA. 117(9):4693-4700.
-
Bacteria self-organize to drive material transport in colonies

Bacteria normally rely on diffusion for material transport, such as nutrient uptake or signal exchange. Diffusion is slow if the distance is long. Many bacterial species use rotating flagella for locomotion. While they move, flagellated bacteria stir their fluidic environment and generate microscale flows. An interesting question is, can bacteria make use of the flagellum-driven flows for long-distance transport? Here we show that flagellated bacteria in colonies can achieve this goal via self-organization. This new mechanism of long-range active transport may bring profound effect on the physiology of bacterial communities in structured environments with heterogeneous nutrient distribution, such as those commonly found in natural and clinical settings. The finding also highlights the physiological significance of phenotypic variability in microbial communities. See [Movie 1], [Movie 2], [Movie 3].
References:
- Haoran Xu, Justas Dauparas, Debasish Das, Eric Lauga, Yilin Wu (2019) Self-organization of swimmers drives long-range fluid transport in bacterial colonies. Nature Communications. 10, 1792.
Past research (before joining CUHK):
-
Osmotic flows drive the spreading of bacterial swarms
When grown on moist surfaces, many flagellated bacteria are able to form a densely packed colony in which millions or more cells move across the surface in a manner called swarming; see this Movie. The swarming colony is enclosed by a thin layer of fluid produced by cells themselves, which provide the milieu to support cell motility. The spreading of swarm fluid is critical to bacterial colonization. Our flow measurements and modeling suggest that osmotic flows sustain the spreading of swarm fluid and fuel the expansion of swarming colonies.
References:
- Wu Y & Berg, H. C. (2012) Water reservoir maintained by cell growth fuels the spreading of a bacterial swarm. PNAS 109(11), 4128–4133.
- Ping L, Wu Y, Hosu BG, Tang JX, Berg HC (2014) Osmotic pressure inside a bacterial swarm. Biophysical Journal 107:871-878.
-
Fluid flows in bacterial swarms
Swarms of flagellated bacteria are swimming within a thin layer of fluid. The swarm fluid is essential for the operation of flagellar motility and for the transport of nutrients or chemical signals. Swarm fluid is only a few microns in thickness, posing great challenges to probe its motion. We found a unique way to generate microscopic bubbles and used these bubbles as tracers for thin-film flows. Using these bubble tracers we discovered a micro-scale river running clockwise (when viewed from above) around the edge of bacterial swarms, which may provide an avenue for long range communication.
References:
- Wu Y, Hosu BG, Berg HC (2011) Microbubbles reveal chiral fluid flows in bacterial swarms. PNAS 108(10): 4147-51.
-
Collective motion of myxobacteria
Collective motion is ubiquitous in biological systems. It is of interest to physics and engineering. Despite the vast differences in length scale and propulsion mechanism, collective motion across biological systems shares a similar feature: Global order arises spontaneously from local interactions between individuals that do not have access to global information. To understand how the traits of individuals give rise to the emergent behavior at population level, we turned to microorganisms because they have a simple behavioral repertoire. We chose to study the collective motion of myxobacteria (Myxococcus xanthus), a well characterized soil bacterium that forms beautiful fruiting bodies. Our modeling results suggested that myxobacteria evolved an unusual behavior to organize efficiently in a crowded environment. The cells reverse moving directions regularly, which facilitates local orientational ordering. We predicted an optimal reversal frequency for this orientational ordering in agreement with the experimental value.
References:
- Wu Y, Jiang Y, Kaiser D, Alber M (2007) Social interactions in myxobacterial swarming. PLoS Comput Biol 3(12): e253.
- Wu Y, Kaiser D, Jiang Y, Alber M (2009) Periodic reversal of direction allows myxobacteria to swarm. Proc Natl Acad Sci USA 106(4): 1222–1227.